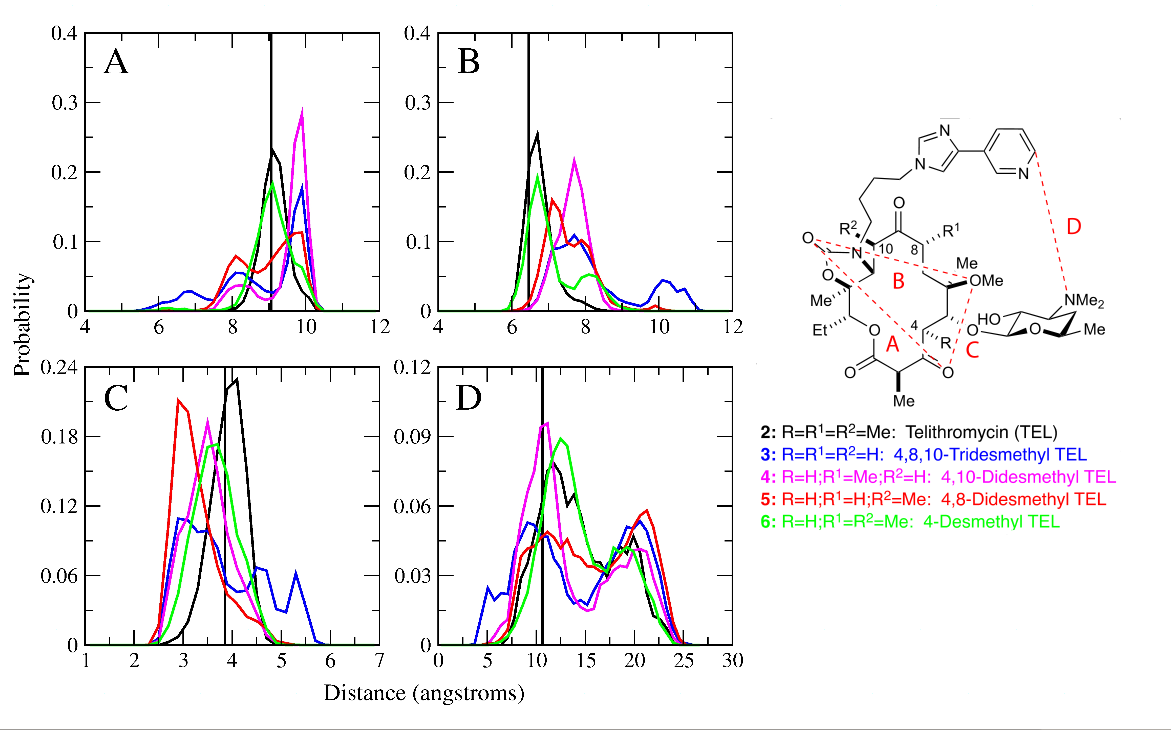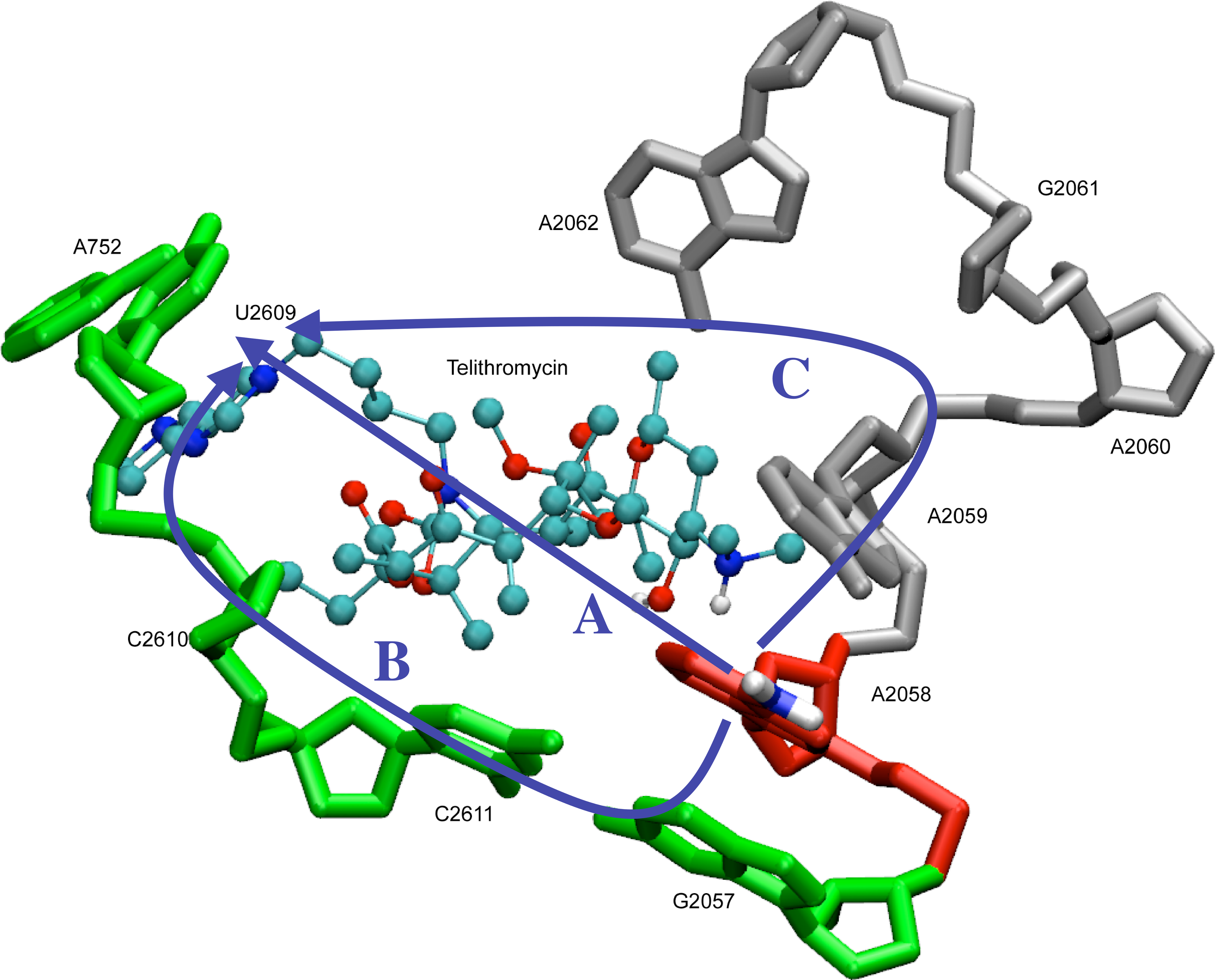
Bacterial resistance to antibiotics is a serious public health problem that requires the continuous development of new antibiotics. Bacteria acquire resistance to macrolide antibiotics by (1) effluxing the drug from the cell, (2) modifying the drug, or (3) modifying the drug target (i.e., the 50S subunit of the ribosome) to abrogate or completely abolish binding. While newer antibiotics are able to avoid the first two mechanisms, they remain unable to overcome resistance due to ribosomal modification, particularly due to methyltransferase (i.e., erm) enzymes. We have applied computer-aided drug design methods designed explicitly for studies of the ribosome to better understand the relationship between modification of the ribosome by erms and the binding of telithromycin, a 3rd generation ketolide antibiotic derived from erythromycin. While we confirm that ribosomal modification leads to decreased binding due to disruption of key interactions with the drug, we find these modifications effect a structural rearrangement of the entire region of the ribosome responsible for binding macrolide antibiotics. This information will be useful in the design of novel antibiotics that are effective against resistant bacteria possessing modified ribosomes.
Article:[PLoS Computational Biology]