Article:[JCC]
| Research Interests molecular modeling, mechanics and dynamics of biological macromolecules and macromolecular assemblies, drug design method development, drug design, force field development |
| Protein Systems studied kinesin motors, tropomyosin, metabotropic glutamate receptors, g-protein coupled-receptors, nuclear receptors, t4-lysozyme |
| Automatic Identification of Selective Conformational Changes in Molecular Simulations |

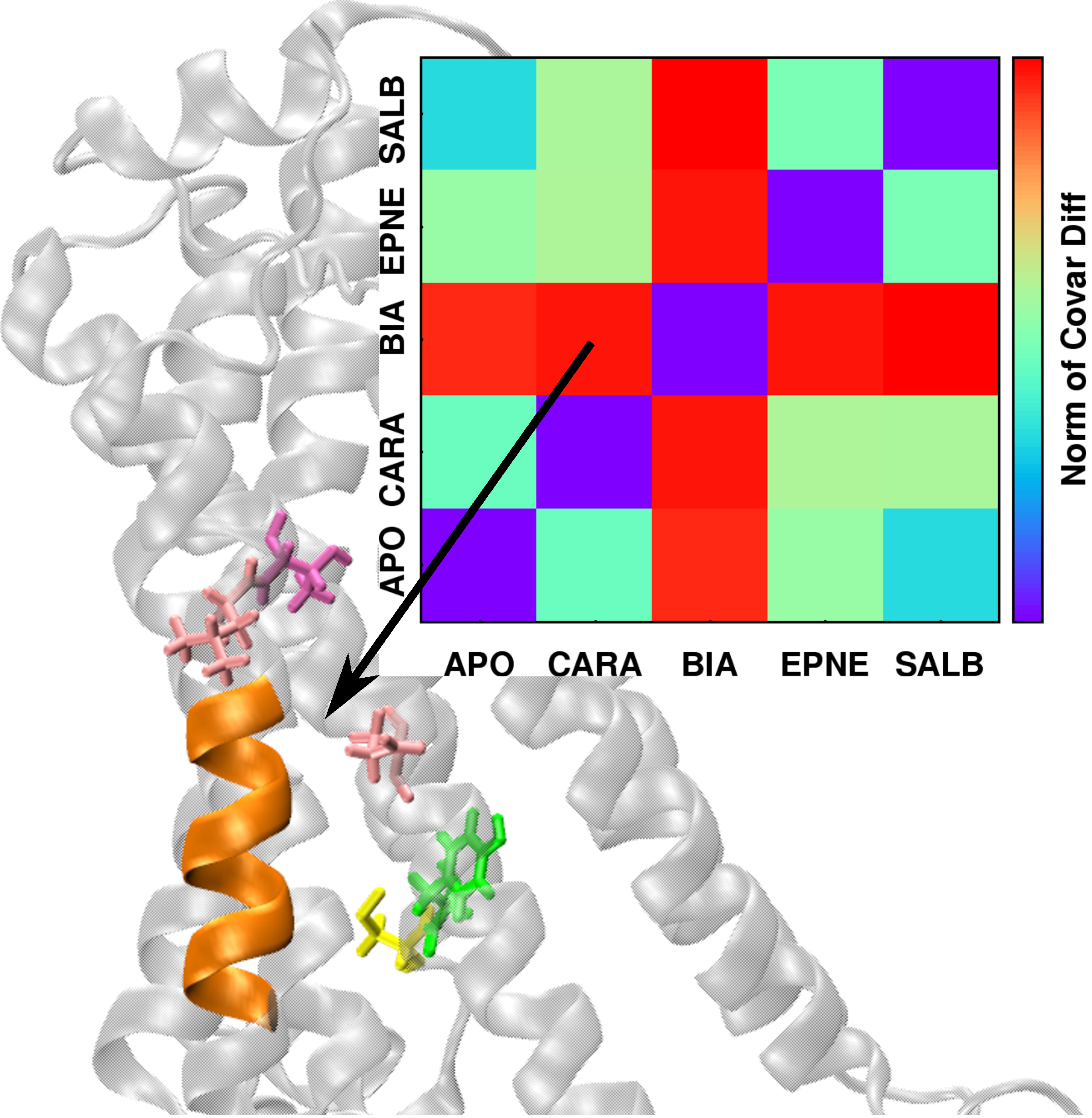
| The conformational dynamics of a macromolecule can be modulated by a number of factors, including changes in environment, ligand binding, and interactions with other macromolecules, among others. We present a method that quantifies the differences in macromolecular conformational dynamics and automatically extracts the structural features that selectively respond to these changes. As a demonstration of its applicability to biomacromolecular systems, the method, referred to as DIRECT-ID, was used to identify relevant ligand-modulated structural variations in the β2-adrenergic (β2AR) G-protein coupled receptor.Micro-second MD simulations of the β2AR in an explicit lipid bilayer were run in the apo state and complexed with the ligands: BI-167107 (agonist), epinephrine (agonist), salbutamol (long-acting partial agonist), or carazolol (inverse agonist). Each ligand modulated the conformational dynamics of β2AR differently and DIRECT-ID analysis of the inverse-agonist vs. agonist-modulated β2AR identified residues known through previous studies to selectively propagate deactivation/activation information, along with some previously unidentified ligand-specific microswitches across the GPCR. This study demonstrates the utility of DIRECT-ID to rapidly extract functionally relevant conformational dynamics information from extended MD simulations of large and complex macromolecular systems. Article:[JCC] |
| Heterogeneous Solute Grand-Canonical Monte Carlo- Molecular Dynamics |

|
Solute sampling of explicit bulk-phase aqueous environments in grand canonical (GC) ensemble simulations suffer from poor convergence due to low insertion probabilities of the solutes. To address this, we developed an iterative procedure involving Grand Canonical-like Monte Carlo (GCMC) and molecular dynamics (MD) simulations. Each iteration involves GCMC of both the solutes and water followed by MD, with the excess chemical potential (μex) of both the solute and the water oscillated to attain their target concentrations in the simulation system. By periodically varying the μex of the water and solutes over the GCMC-MD iterations, solute exchange probabilities and the spatial distributions of the solutes improved. The utility of the oscillating-μex GCMC-MD method is indicated by its ability to approximate the hydration free energy (HFE) of the individual solutes in aqueous solution as well as in dilute aqueous mixtures of multiple solutes. For seven organic solutes: benzene, propane, acetaldehyde, methanol, formamide, acetate, and methylammonium, the average μex of the solutes and the water converged close to their respective HFEs in both 1 M standard state and dilute aqueous mixture systems. The oscillating-μex GCMC methodology is also able to drive solute sampling in proteins in aqueous environments as shown using the occluded binding pocket of the T4 lysozyme L99A mutant as a model system. The approach was shown to satisfactorily reproduce the free energy of binding of benzene as well as sample the functional group requirements of the occluded pocket consistent with the crystal structures of known ligands bound to the L99A mutant as well as their relative binding affinities. Article:[JCTC] |
| Estimation of ligand efficacies of metabotropic glutamate receptors from conformational forces obtained from molecular dynamics simulations |
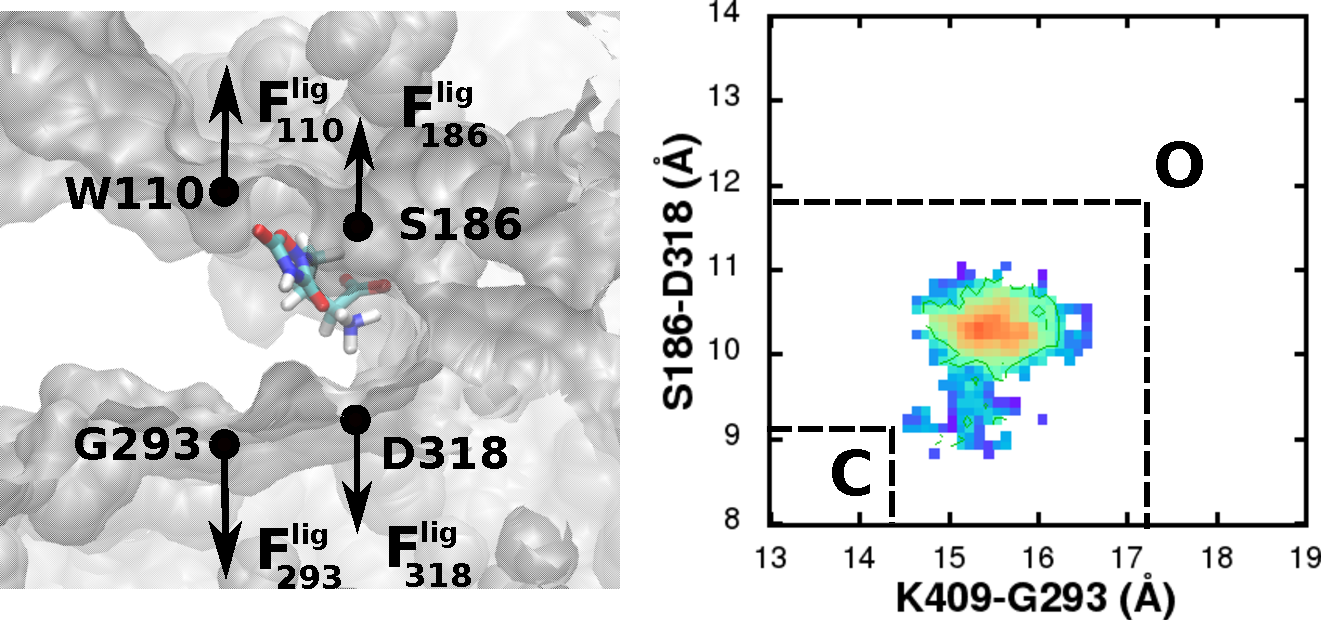 |
Group 1 metabotropic glutamate receptors (mGluR) are G-protein coupled receptors
with a large bilobate extracellular ligand binding region (LBR) that resembles a Venus fly trap.
Closing of this LBR in the presence of a ligand is associated with the activation of the receptor.
From conformational sampling of the LBR-ligand complexes using all-atom molecular dynamics (MD) simulations,
we characterized the conformational minima related to the hinge like motion associated with the LBR closing/opening
in the presence of known agonists and antagonists.
By applying a harmonic restraint on the LBR, we also determined the conformational forces generated by the different ligands.
The change in the location of the minima and the conformational forces were used to quantify the efficacies of the ligands.
This analysis shows that efficacies can be estimated from the forces of a single conformation of the receptor,
indicating the potential of MD simulations as an efficient and useful technique to quantify efficacies,
thereby facilitating the rational design of mGluR agonists and antagonists. |
| Previous Research at the Molecular Biomechanics Lab, Texas A&M University |
| Hyteresis-based mechanism for the directed motility of the Ncd motor |
|
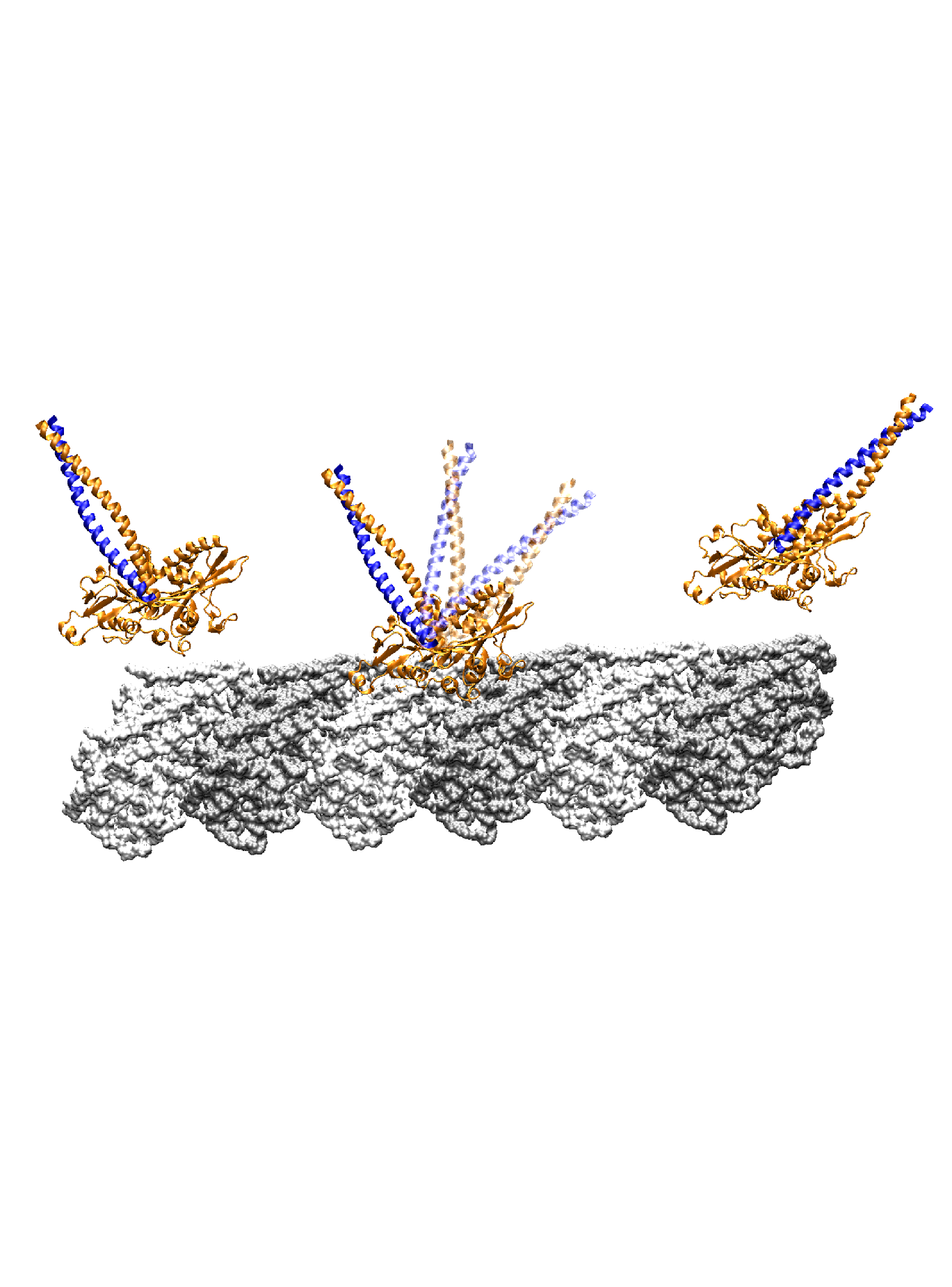
Ncd is a homodimer of the kinesin superfamily, involved in microtubule organization in spindle assembly during mitosis and meiosis. Despite having a motor head domain similar to Kin, Ncd is non-processive and has a minus-end directed movement on microtubules. Walking step of Ncd involves a lever arm like rotation of its neck to the minus-end. Using a combination of molecular dynamics simulations and structural analyses, we find that the neck sequentially makes intermediate contacts with the motor head along its mostly longitudinal path, and it develops a 24° twist in the post-stroke orientation. The forward (pre-stroke to post-stroke) motion has an 4.5 kBT (where kB is the Boltzmann constant, and T = 300 K) free-energy barrier and is a diffusion guided by the intermediate contacts. The post-stroke free-energy minimum is higher and is formed ±10° before reaching the orientation in the post-stroke crystal structure, consistent with previous structural data. The importance of intermediate contacts correlates with the existing motility data, including those for mutant Ncds. Unlike the forward motion, the recovery stroke goes nearly downhill in free energy, powered in part by torsional relaxation of the neck. The hysteresis in the energetics of the neck motion arises from the mechanical compliance of the protein, and together with guided diffusion, it may be key to the directed motility of Ncd. Article:[Biophys J] |
| Functionally distinct regions of tropomyosin have different flexibilities |
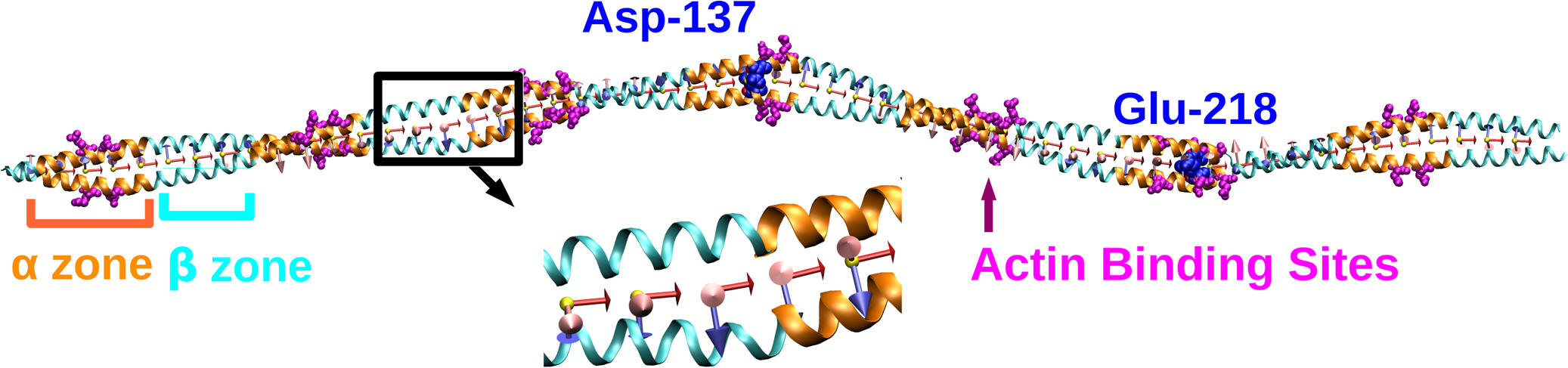
|
Tropomyosin makes up a large complex family of
alpha helical coiled-coils found in eukaryotes, and is usually seen bound to F-actin.
In thin filaments of muscle, it regulates actin-myosin interactions by winding around actin in a
right handed fashion and blocking the actin binding sites of myosin.
While precise roles of nonmuscle tropomyosin isoforms such as those found in
stress fiber assemblies is poorly understood, tropomyosin has been observed to increase actin's persistence length by about 1.5 times.
It could thus significantly affect mechanical properties of the cytoskeleton.
Persistence length of tropomyosin has previously been measured to be about 150 nm
(Hvidt, S. et al., 1982. Biochem. 21, 4064).
However, recent studies show that long coiled coils such as the stalk of
a Rad50 complex exhibit regional variations in flexibilities (van Noort, J. et al., 2003. PNAS, 100, 7581).
|
| Conformation of alpha helices, coiled coils and biofilaments is governed by their Critical Buckling Length |
|
Supra molecular assemblies of
commonly found secondary structures of proteins called α-helices have varied mechanical roles in a sub-cellular environment.
Alpha helices are wound in a right-handed fashion due to a hydrogen bonding between the C=O and the N-H atoms
across every i and i+4th residue in the poly-peptide chain.
It has long been established that this backbone hydrogen bonding form the elastic core of α-helices,
keeping the flexural rigidity independent of the amino acid sequence composition.
Several characterizations of their mechanical properties using either experimental or computational techniques,
hence treated α-helices as homogenous and linear elastic rods and their stiffness typically expressed as persistence length.
We find that bending stiffness of α-helices, coiled coils, and biofilaments in general,
depends on its length, due to inherent non-bonded attractions that are typically not screened in physiological conditions.
In particular, non-bonded attraction introduces a length scale,
critical buckling length, beyond which a biofilament can no longer remain linearly elastic.
These results suggest that non-bonded attractions can significantly affect elastic properties of biofilament systems such as the cytoskeleton. |